Fabian Heinemann
Toxicity Assessment in Preclinical Histopathology via Class-Aware Mahalanobis Distance for Known and Novel Anomalies
Feb 02, 2026Abstract:Drug-induced toxicity remains a leading cause of failure in preclinical development and early clinical trials. Detecting adverse effects at an early stage is critical to reduce attrition and accelerate the development of safe medicines. Histopathological evaluation remains the gold standard for toxicity assessment, but it relies heavily on expert pathologists, creating a bottleneck for large-scale screening. To address this challenge, we introduce an AI-based anomaly detection framework for histopathological whole-slide images (WSIs) in rodent livers from toxicology studies. The system identifies healthy tissue and known pathologies (anomalies) for which training data is available. In addition, it can detect rare pathologies without training data as out-of-distribution (OOD) findings. We generate a novel dataset of pixelwise annotations of healthy tissue and known pathologies and use this data to fine-tune a pre-trained Vision Transformer (DINOv2) via Low-Rank Adaptation (LoRA) in order to do tissue segmentation. Finally, we extract features for OOD detection using the Mahalanobis distance. To better account for class-dependent variability in histological data, we propose the use of class-specific thresholds. We optimize the thresholds using the mean of the false negative and false positive rates, resulting in only 0.16\% of pathological tissue classified as healthy and 0.35\% of healthy tissue classified as pathological. Applied to mouse liver WSIs with known toxicological findings, the framework accurately detects anomalies, including rare OOD morphologies. This work demonstrates the potential of AI-driven histopathology to support preclinical workflows, reduce late-stage failures, and improve efficiency in drug development.
A comparative evaluation of image-to-image translation methods for stain transfer in histopathology
Apr 06, 2023



Abstract:Image-to-image translation (I2I) methods allow the generation of artificial images that share the content of the original image but have a different style. With the advances in Generative Adversarial Networks (GANs)-based methods, I2I methods enabled the generation of artificial images that are indistinguishable from natural images. Recently, I2I methods were also employed in histopathology for generating artificial images of in silico stained tissues from a different type of staining. We refer to this process as stain transfer. The number of I2I variants is constantly increasing, which makes a well justified choice of the most suitable I2I methods for stain transfer challenging. In our work, we compare twelve stain transfer approaches, three of which are based on traditional and nine on GAN-based image processing methods. The analysis relies on complementary quantitative measures for the quality of image translation, the assessment of the suitability for deep learning-based tissue grading, and the visual evaluation by pathologists. Our study highlights the strengths and weaknesses of the stain transfer approaches, thereby allowing a rational choice of the underlying I2I algorithms. Code, data, and trained models for stain transfer between H&E and Masson's Trichrome staining will be made available online.
Learning image representations for anomaly detection: application to discovery of histological alterations in drug development
Oct 14, 2022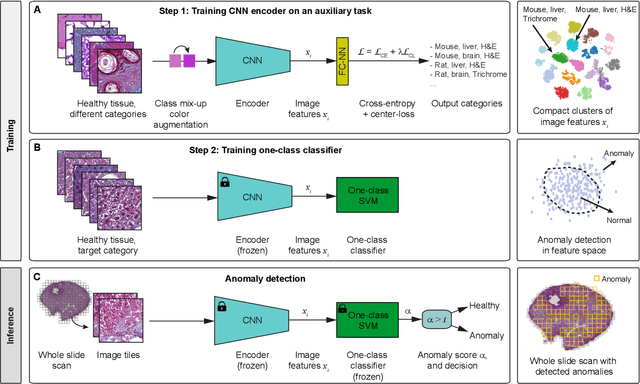
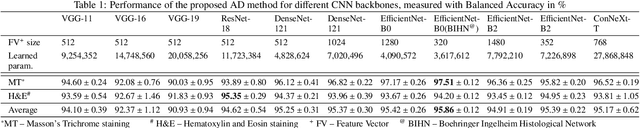
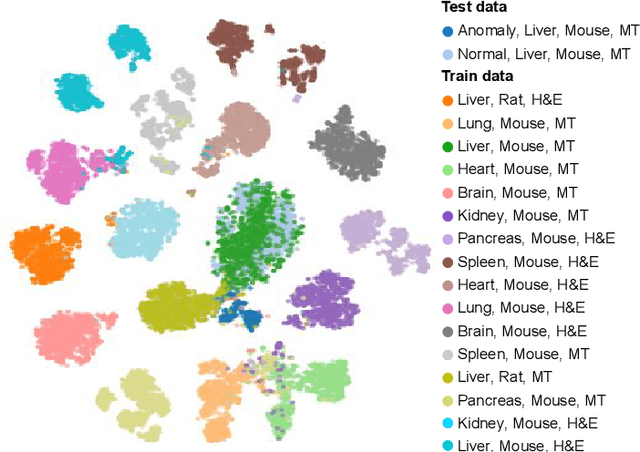
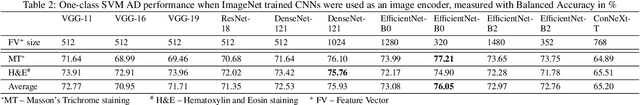
Abstract:We present a system for anomaly detection in histopathological images. In histology, normal samples are usually abundant, whereas anomalous (pathological) cases are scarce or not available. Under such settings, one-class classifiers trained on healthy data can detect out-of-distribution anomalous samples. Such approaches combined with pre-trained Convolutional Neural Network (CNN) representations of images were previously employed for anomaly detection (AD). However, pre-trained off-the-shelf CNN representations may not be sensitive to abnormal conditions in tissues, while natural variations of healthy tissue may result in distant representations. To adapt representations to relevant details in healthy tissue we propose training a CNN on an auxiliary task that discriminates healthy tissue of different species, organs, and staining reagents. Almost no additional labeling workload is required, since healthy samples come automatically with aforementioned labels. During training we enforce compact image representations with a center-loss term, which further improves representations for AD. The proposed system outperforms established AD methods on a published dataset of liver anomalies. Moreover, it provided comparable results to conventional methods specifically tailored for quantification of liver anomalies. We show that our approach can be used for toxicity assessment of candidate drugs at early development stages and thereby may reduce expensive late-stage drug attrition.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge